Within the RT Module (but not the CT Module), GoldSim provides an option to utilize a large built-in database of nuclide decay data (species, decay rates and daughter products). In particular, the feature allows you to view over 1200 nuclides (including both stable and radioactive species) and to selectively include some of these species in your simulation.
The following should be noted regarding the data used by GoldSim:
•The data is based on the International Commission for Radiation Protection (ICRP)’s Publication 107 (data table ICRP107.NDX). This reference can be viewed here: http://www.icrp.org/publication.asp?id=ICRP Publication 107)
•The ICRP table of nuclides contains a list of nuclides with up to three daughters each, along with daughter stoichiometry (i.e., decay branching fractions) and atomic weights.
•The GoldSim implementation uses an updated data set provided by Dr. Keith Eckerman of Oak Ridge National Laboratory in February 2012. This data set corrected minor errors in the atomic weights of some isotopes.
•In some cases where nuclide decays into multiple daughter products, the sum of the ICRP data set’s daughter stoichiometry factor is slightly greater than 1.0 due to roundoff. In these cases, GoldSim reduces the stoichiometry factor for the commonest daughter so the sum of all stoichiometry factors equals 1.0.
•There are some instances where daughter products are not fully specified in the ICRP data set. This is the case for some heavy nuclides (e.g., isotopes of Cf and Cm) that are subject to spontaneous fission, and for some fractional daughters of other nuclides.
•The stable elements included in the data set use the average atomic weight for the naturally-occurring isotopes of the element. This implies that the product of the last decay in a given chain joins the ‘pool’ of naturally-occurring isotopes of the final element.
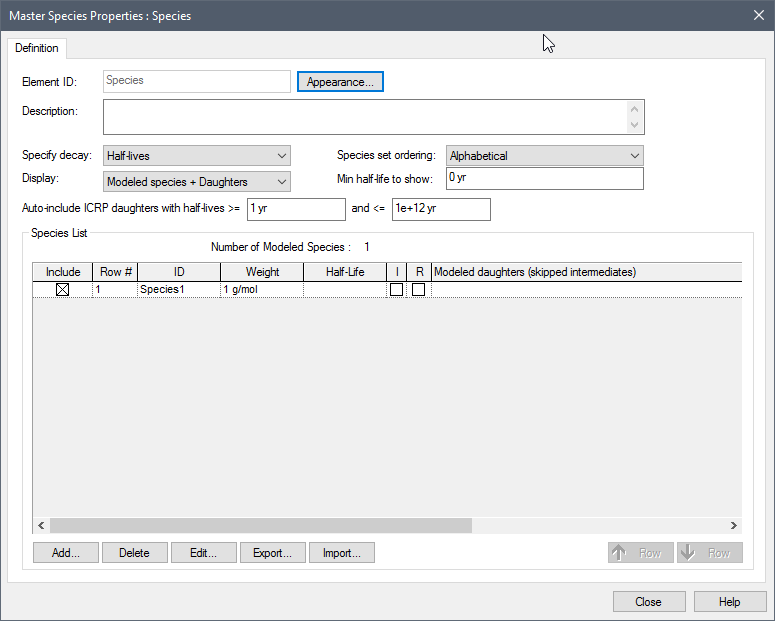
Within the RT Module, when you open the Species dialog for the first time, it looks like this:

The Display field provides access to the full ICRP database. This drop list provides three options:
•Modeled species (the default): Displays only those species for which the Include box is checked.
•Modeled species + Daughters: Displays only those species for which the Include box is checked and all of their progeny.
•All species: Displays all species.
 Note: If you are using the RT
Module but do not wish to access species from the ICRP database (and just
want to add ‘user-defined” species yourself), simply make sure that the
Display field remains set to the default (“Modeled species”).
Note: If you are using the RT
Module but do not wish to access species from the ICRP database (and just
want to add ‘user-defined” species yourself), simply make sure that the
Display field remains set to the default (“Modeled species”).
By default, Display is set to “Modeled species” and a single “user-defined” species (Species1) is listed (and the Include box is checked).
 Note: The Include box
is always checked (and cannot be cleared) for any “user-defined” species (the
default species initially present in all new models, and any additional species
added via the Add button).
Note: The Include box
is always checked (and cannot be cleared) for any “user-defined” species (the
default species initially present in all new models, and any additional species
added via the Add button).
To utilize the ICRP database, you will typically initially select “All species”. When you do so, the Min half-life to show field becomes available. GoldSim will then display all species from the database (and any custom species you have added manually) whose half-life is greater than that specified or whose Include box is checked. That is, GoldSim will exclude from display species which are not Included and whose half-life is less than the specified value.
Although GoldSim will display a large number of species when you select “All species”, it is important to understand that this does not mean that they will all be included in your model. The Include checkbox is what defines the species to be modeled. That is, only species for which the Include box is checked are included in the model. Checking the Include box for any species includes it in the model, and clearing the box excludes it from the model.
The final column of the Species element, Modeled daughters (skipped intermediates), indicates which daughters are included in the simulation (and which are excluded), and this is important information to note. If a daughter (or other descendant) of the species is included, it is listed without parentheses. If it is not included, it is listed surrounded by parentheses.
Although the ICRP database includes a very large number of species, any particular model will only use a small subset of these. The subset you select to use will likely be a function of the inventory present, the length of the simulation, and the time scale of the processes being simulated. In most cases, the half-lives of the various species (and their daughters) will play a key role in determining which species are to be included in the model. To facilitate this, GoldSim provides an option to Auto-include ICRP daughters within a specified range of half-lives.
To illustrate how the various options can be used, it is most instructive to outline the steps that you will typically take to create your modeled species list using the ICRP database:
1. Select “All species” from the Display field.
2. Depending on the time frame of your simulation and the time scale of the processes being simulated, you may then want to specify a Min half-life to show in order to hide species with very short half-lives that you know you will not explicitly include in your simulation.
3. In most cases, you will have a list of key species you know must be included in the simulation (e.g., for a disposal facility, species you know will be in the disposal inventory). You should check the Include box for each species. Note that you need not consider the daughters for these key species yet. As will be seen in the next step, GoldSim provides a method for automatically including these.
 Note: GoldSim provides some
tools to facilitate finding and selecting species for inclusion. In particular,
in any column in the Species dialog, clicking on it sorts it (in ascending
order). Clicking it again sorts it in descending order. Clicking a
column and then typing in a key combination takes you immediately to that row
(e.g., typing Th would take you to the first Thorium isotope in the
list).
Note: GoldSim provides some
tools to facilitate finding and selecting species for inclusion. In particular,
in any column in the Species dialog, clicking on it sorts it (in ascending
order). Clicking it again sorts it in descending order. Clicking a
column and then typing in a key combination takes you immediately to that row
(e.g., typing Th would take you to the first Thorium isotope in the
list).
4. If you now select “Modeled species + daughters” from the Display list, GoldSim will show only the key species that you have manually included, as well as all of the daughters (in fact, the full chain of descendants) of the key species. You will note that the Include box for some of these daughters will be selected and grayed out:
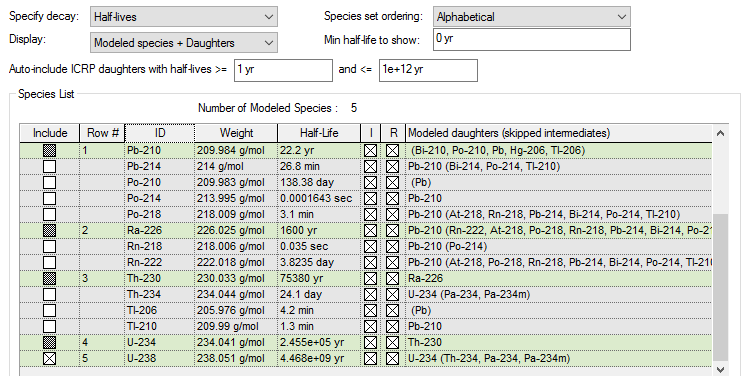
Note that included species are highlighted in green.
In this example, U-238 has been manually included (the Include box has an X). As a result, 4 other nuclides (which are descendants of U-238) have been auto-included (and their Include boxes are marked as gray). Th-234 (and a number of other nuclides) are listed, but excluded (i.e., the Include box is cleared).
The reason for these selections can be explained by the options selected directly above. Since Display is set to “Modeled species + daughters”, the daughters of U-238 (and any other manually selected species) are shown in the list. Because Min half-life to show is set to 0, none of the daughters are hidden; even very short-lived daughters (such as Th-234) are shown. Finally, because the range for Auto-include ICRP daughters is set from 1 yr to 1e+12 years, only daughters with half-lives in that range are auto-included.
Hence, auto-inclusion provides a quick and convenient way to include important daughters in the model without having to do so manually.
 Note: You cannot exclude
(clear the Include box) for an auto-included species. It is always
included, unless the parent that caused it to be auto-included is manually
excluded).
Note: You cannot exclude
(clear the Include box) for an auto-included species. It is always
included, unless the parent that caused it to be auto-included is manually
excluded).
5. Based on your auto-inclusion settings, some daughters may still be excluded. You may want to examine these to determine if you would like to include them for a special reason. For example, stable species at the end of the chain will not be auto-included, and you may want to include these if you think they may be present in significant quantities. Also, you may want to include short-lived species (e.g., Rn-222) that have special transport properties (e.g., they are volatile).
6. All new models initially have a “user-defined” species called Species1. You cannot delete this until you have selected other species to be included (there must always be at least one species). If you have not already done so, you should delete this (by selecting the row and pressing the Delete button).
7. To simplify the display, you may want to set the Display setting to “Modeled species” such that only the species included in the simulation are shown.
8. You may want to set the Species set ordering to define the order of species in the Species array label set.
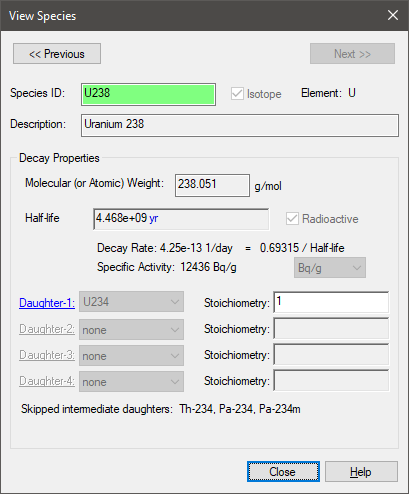
It is important to understand what it means to exclude a daughter in a chain. When we do so, the parents of those species “skip over” them and proceed to daughters that have been included. To understand this, consider the U-238 decay chain. The first part of the chain actually looks like this:
U-238 => Th-234 => Pa-234m => U-234 …
However, Th-234 and Pa-234m have short half-lives (less than 1 yr), so if we exclude those two species, GoldSim “skips” them in the decay chain such that U-238 is assumed to decay directly to U-234:
Several additional points should be noted regarding the use of ICRP species:
•Although the ICRP species are shown (in the ID column) with a hyphen separating the element name from the mass number, within the corresponding Species array label set, the hyphen is eliminated (hence, U-238 becomes U238 in the species array label set).
•GoldSim will not let you create a “user-defined” species whose name conflicts with an included species from the ICRP database. Hence, if you try to create a “user-defined” species called U238 and the ICRP species U-238 is marked as included, GoldSim will not let you create the species. Similarly, if a “user-defined” species named U238 already exists, you will not be able to mark the ICRP species U-238 as included.
•You cannot edit the properties of the ICRP species. If you click on them in the Species dialog, a dialog for that species will be displayed, but none of the properties can be edited. The Species ID of an ICRP species is highlighted in green:
•GoldSim provides the ability to export your species list and subsequently import it (e.g., into another model). If you export a list of ICRP species, when they are subsequently imported, they are always treated as “user-defined” species.
Learn more about: